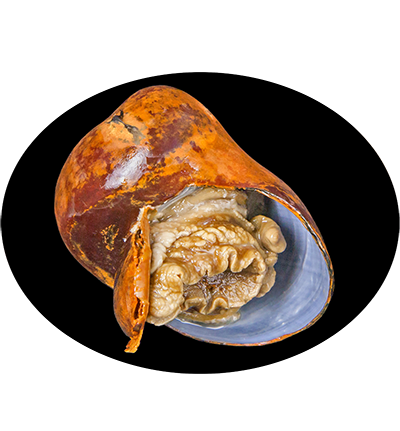
Gigantopelta aegis is a species of deep-sea snail in the family Peltospiridae, found in hydrothermal vent ecosystems, particularly in the Longqi hydrothermal vent field on the Southwest Indian Ridge. This species is adapted to extreme environments, relying on a symbiotic relationship with chemosynthetic bacteria housed in its gills. These bacteria oxidize sulfur compounds from vent fluids to produce organic matter, providing essential nutrients to the snail. Gigantopelta aegis plays a crucial role in vent ecosystems by contributing to nutrient cycling and supporting microbial and faunal communities.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Gigantopelta aegis | Mollusca | deep-sea hydrothermal vent snail | Hydrothermal vent | >2,761 | Longqi hydrothermal vent field (37.7839°S, 49.6502°E) on the Southwest Indian Ridge | 1735272 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| Gae_host_genome | 1.1Gb | Chromosome | 2021 | JAEHGF01 | Hong Kong University of Science and Technology | PRJNA612619 | 94.60% | 22,556 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Hologenome analysis reveals dual symbiosis in the deep-sea hydrothermal vent snail Gigantopelta aegis | nature communications | 33608555 |
| Gene ID | Description |
|---|---|
| ctg4387-0.1-1 | TYROSINE-PROTEIN KINASE RECEPTOR |
| ctg4387-0.5-1 | |
| ctg439-0.2-1 | CYTOCHROME P450 508A4-RELATED |
| ctg439-1.2-1 | POU DOMAIN |
| ctg439-2.7-1 | UDP-GLUCOSE DEHYDROGENASE/UDP-MANNAC DEHYDROGENASE |
| ctg439-3.20-1 | TRANSLATION INITIATION FACTOR EIF-2B SUBUNIT EPSILON |
| ctg439-3.21-1 | L-ASPARAGINASE |
| ctg439-3.22-1 | |
| ctg439-3.6-1 | MYELIN PROTEOLIPID |
| ctg439-3.9-1 | PUTATIVE-RELATED |
| ctg4390-0.0-1 | E3 UBIQUITIN-PROTEIN LIGASE NHLRC1-RELATED |
| ctg4391-0.0-1 | TRANSCRIPTION INITIATION FACTOR TFIID |
| ctg4393-0.5-1 | MACROGLOBULIN / COMPLEMENT |
| ctg4393-0.6-1 | MACROGLOBULIN / COMPLEMENT |
| ctg4396-0.3-1 | |
| ctg4396-0.4-1 | MANNOSYL-OLIGOSACCHARIDE GLUCOSIDASE |
| ctg4397-0.3-1 | ZGC:195170 |
| ctg4397-0.6-1 | SI:CH211-108C17.2-RELATED-RELATED |
| ctg44-0.1-1 | INHIBITOR OF APOPTOSIS |
| ctg44-0.12-1 | |
| ctg44-0.2-1 | INTEGRASE CORE DOMAIN CONTAINING PROTEIN |
| ctg44-0.3-1 | |
| ctg44-1.0-1 | LEUCINE AMINOPEPTIDASE-RELATED |
| ctg44-1.1-1 | |
| ctg44-1.2-1 | ANAPHASE-PROMOTING COMPLEX SUBUNIT 5 |
| ctg44-1.3-1 | DNA-DIRECTED RNA POLYMERASE I SUBUNIT 2 |
| ctg44-2.0-1 | RIKEN CDNA 2310061I04 GENE |
| ctg44-2.1-1 | PROTEASE-ASSOCIATED DOMAIN-CONTAINING PROTEIN |
| ctg44-2.2-1 | ER DEGRADATION-ENHANCING ALPHA-MANNOSIDASE-LIKE PROTEIN 2 |
| ctg44-2.4-1 | DUAL SPECIFICITY PROTEIN PHOSPHATASE 12 FAMILY MEMBER |
| ctg44-3.0-1 | HOMOSERINE O-ACETYLTRANSFERASE |
| ctg44-3.2-1 | RIBONUCLEASE III |
| ctg44-3.3-1 | 60 KDA RIBONUCLEOPROTEIN SSA/RO |
| ctg44-3.4-1 | VACUOLAR ATP SYNTHASE SUBUNIT G |
| ctg44-3.5-1 | PROTEIN ABHD14B-LIKE |
| ctg44-4.5-1 | GOLGI SNARE BET1-RELATED |
| ctg44-4.6-1 | ELONGATION FACTOR TU-RELATED |
| ctg44-4.7-1 | ARGININOSUCCINATE SYNTHASE |
| ctg44-5.10-1 | PHYTANOYL-COA DIOXYGENASE DOMAIN CONTAINING 1 |
| ctg44-5.11-1 | B-CELL LYMPHOMA/LEUKEMIA 10 |
| ctg44-5.12-1 | |
| ctg44-5.13-1 | SJOEGREN SYNDROME NUCLEAR AUTOANTIGEN 1 |
| ctg44-6.0-1 | SH3 MULTIPLE DOMAIN |
| ctg44-7.0-1 | PROMININ PROM PROTEIN |
| ctg44-7.1-1 | RHO FAMILY GTPASE |
| ctg44-7.6-1 | ENZYMATIC POLYPROTEIN-RELATED |
| ctg44-8.6-1 | PROMININ PROM PROTEIN |
| ctg44-8.7-1 | |
| ctg44-9.15-1 | PROMININ PROM PROTEIN |
| ctg44-9.16-1 | PROMININ PROM PROTEIN |

