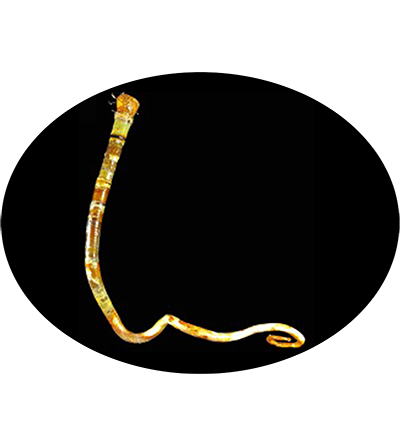
The vestimentiferan Paraescarpia echinospica is widely distributed in the cold seeps and methane seeps of the western Pacific Ocean, and relies on their endosymbiont bacteria as a source of energy and organic carbon. It's gutless and depends entirely on its endosymbiotic sulfide-oxidizing chemoautotrophic bacteria for nutrition. Its mechanisms of host–symbiont cooperation in energy production and nutrient biosynthesis and utilization have recently been documented through a study of its endosymbiont genome and metaproteome.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Paraescarpia echinospica | Annelida | deep-sea tubeworm | Cold seep/Hydrothermal vent | 1100-1600 | Haima cold seep in the South China Sea (16°43.80′N, 110°28.50′E) | 2080241 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| HKBU_Pec_v1 | 1.09Gb | Chromosome | 2021 | JAHLWY01 | Hong Kong University of Science and Technology | PRJNA625616 | 96.40% | 22,642 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic Signatures Supporting the Symbiosis and Formation of Chitinous Tube in the Deep-Sea Tubeworm Paraescarpia echinospica | Molecular Biology and Evolution | 34255082 |
| Gene ID | Description |
|---|---|
| PE_Scaf960_1.4 | SI:CH211-108C17.2-RELATED-RELATED |
| PE_Scaf960_10.3 | |
| PE_Scaf960_6.4 | DELTEX-RELATED |
| PE_Scaf960_7.3 | PARP/ZINC FINGER CCCH TYPE DOMAIN CONTAINING PROTEIN |
| PE_Scaf9617_1.3 | ZINC FINGER, C3HC4 TYPE (RING FINGER) FAMILY PROTEIN |
| PE_Scaf9617_3.3 | PROTEIN FAM72A-RELATED |
| PE_Scaf9617_5.10 | ACETYLCHOLINESTERASE |
| PE_Scaf9617_5.19 | SLIT-ROBO RHO GTPASE ACTIVATING PROTEIN |
| PE_Scaf9617_5.20 | SLIT-ROBO RHO GTPASE ACTIVATING PROTEIN |
| PE_Scaf9617_5.21 | SLIT-ROBO RHO GTPASE ACTIVATING PROTEIN |
| PE_Scaf9617_5.22 | - |
| PE_Scaf9617_6.16 | UNCHARACTERIZED |
| PE_Scaf9617_6.7 | PROTEASE M14 CARBOXYPEPTIDASE |
| PE_Scaf9617_6.9 | ACETYLCHOLINESTERASE |
| PE_Scaf9618_0.0 | |
| PE_Scaf9619_2.12 | AFADIN |
| PE_Scaf9619_2.8 | INTRAFLAGELLAR TRANSPORT PROTEIN 88 HOMOLOG |
| PE_Scaf961_0.3 | |
| PE_Scaf9620_11.0 | |
| PE_Scaf9620_2.2 | |
| PE_Scaf9620_5.4 | AB HYDROLASE SUPERFAMILY PROTEIN C4A8.06C |
| PE_Scaf9620_7.4 | AB HYDROLASE SUPERFAMILY PROTEIN C4A8.06C |
| PE_Scaf9620_8.3 | |
| PE_Scaf9622_0.1 | COLLAGEN IV NC1 DOMAIN-CONTAINING PROTEIN |
| PE_Scaf9624_1.3 | GENERAL RECEPTOR FOR PHOSPHOINOSITIDES 1-ASSOCIATED SCAFFOLD PROTEIN-RELATED |
| PE_Scaf9624_2.2 | BARDET-BIEDL SYNDROME 2 PROTEIN |
| PE_Scaf9624_2.3 | ALPHA-CATULIN |
| PE_Scaf9625_1.0 | |
| PE_Scaf9625_1.2 | |
| PE_Scaf9625_2.2 | |
| PE_Scaf9625_3.0 | |
| PE_Scaf9625_3.10 | CYTOCHROME P450 FAMILY 3 |
| PE_Scaf9625_3.11 | |
| PE_Scaf9625_3.8 | |
| PE_Scaf9628_2.0 | |
| PE_Scaf9628_3.0 | |
| PE_Scaf9628_4.4 | YEAST SPT2-RELATED |
| PE_Scaf9628_4.5 | PROTEIN LIN-7 HOMOLOG |
| PE_Scaf9628_4.6 | CHOLINE/ETHANOALAMINE KINASE |
| PE_Scaf9628_5.1 | POTASSIUM CHANNEL TETRAMERIZATION DOMAIN-CONTAINING |
| PE_Scaf9628_5.10 | 60S RIBOSOMAL PROTEIN L27A |
| PE_Scaf9628_5.5 | DENN DOMAIN-CONTAINING PROTEIN 2 |
| PE_Scaf9628_6.0 | |
| PE_Scaf9628_6.3 | FOUR-JOINTED BOX PROTEIN 1 |
| PE_Scaf9628_6.4 | TELOMERIC REPEAT-BINDING FACTOR 1 TERF1 |
| PE_Scaf9629_0.3 | UNCHARACTERIZED |
| PE_Scaf9629_0.4 | SERINE/THREONINE PROTEIN KINASE |
| PE_Scaf9630_0.6 | |
| PE_Scaf9630_0.7 | MANNOSE, PHOSPHOLIPASE, LECTIN RECEPTOR RELATED |
| PE_Scaf9631_0.3 | CADHERIN-87A |

