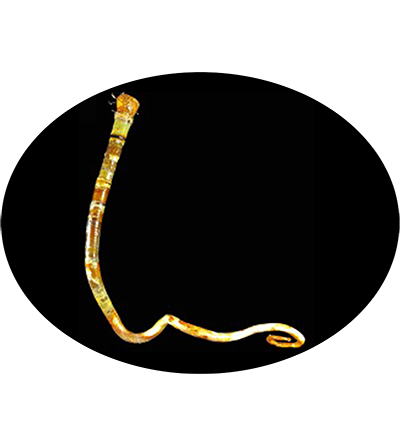
The vestimentiferan Paraescarpia echinospica is widely distributed in the cold seeps and methane seeps of the western Pacific Ocean, and relies on their endosymbiont bacteria as a source of energy and organic carbon. It's gutless and depends entirely on its endosymbiotic sulfide-oxidizing chemoautotrophic bacteria for nutrition. Its mechanisms of host–symbiont cooperation in energy production and nutrient biosynthesis and utilization have recently been documented through a study of its endosymbiont genome and metaproteome.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Paraescarpia echinospica | Annelida | deep-sea tubeworm | Cold seep/Hydrothermal vent | 1100-1600 | Haima cold seep in the South China Sea (16°43.80′N, 110°28.50′E) | 2080241 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| HKBU_Pec_v1 | 1.09Gb | Chromosome | 2021 | JAHLWY01 | Hong Kong University of Science and Technology | PRJNA625616 | 96.40% | 22,642 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic Signatures Supporting the Symbiosis and Formation of Chitinous Tube in the Deep-Sea Tubeworm Paraescarpia echinospica | Molecular Biology and Evolution | 34255082 |

