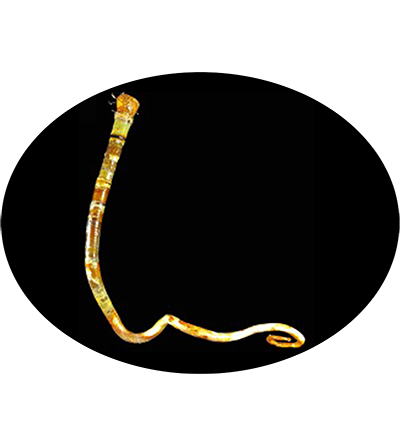
The vestimentiferan Paraescarpia echinospica is widely distributed in the cold seeps and methane seeps of the western Pacific Ocean, and relies on their endosymbiont bacteria as a source of energy and organic carbon. It's gutless and depends entirely on its endosymbiotic sulfide-oxidizing chemoautotrophic bacteria for nutrition. Its mechanisms of host–symbiont cooperation in energy production and nutrient biosynthesis and utilization have recently been documented through a study of its endosymbiont genome and metaproteome.
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Paraescarpia echinospica | Annelida | deep-sea tubeworm | Cold seep/Hydrothermal vent | 1100-1600 | Haima cold seep in the South China Sea (16°43.80′N, 110°28.50′E) | 2080241 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness | Gene Number |
|---|---|---|---|---|---|---|---|---|
| HKBU_Pec_v1 | 1.09Gb | Chromosome | 2021 | JAHLWY01 | Hong Kong University of Science and Technology | PRJNA625616 | 96.40% | 22,642 |
| Title | Journal | Pubmed ID |
|---|---|---|
| Genomic Signatures Supporting the Symbiosis and Formation of Chitinous Tube in the Deep-Sea Tubeworm Paraescarpia echinospica | Molecular Biology and Evolution | 34255082 |
| Gene ID | Description |
|---|---|
| PE_Scaf9722_1.8 | CARBONIC ANHYDRASE |
| PE_Scaf9725_0.5 | DESTABILASE-RELATED |
| PE_Scaf9727_1.3 | RNA (RNA) POLYMERASE II ASSOCIATED PROTEIN HOMOLOG |
| PE_Scaf9727_1.4 | |
| PE_Scaf9727_3.5 | |
| PE_Scaf9727_3.6 | ZINC FINGER PROTEIN |
| PE_Scaf9728_0.0 | |
| PE_Scaf9729_0.3 | CELL DIVISION PROTEIN KINASE |
| PE_Scaf9732_0.7 | OS10G0105400 PROTEIN |
| PE_Scaf9733_1.7 | ZGC:195170 |
| PE_Scaf9733_1.8 | ATP-BINDING CASSETTE SUB-FAMILY C |
| PE_Scaf9736_1.5 | |
| PE_Scaf9736_3.11 | ATP-BINDING CASSETTE SUB-FAMILY C |
| PE_Scaf9736_3.12 | ATP-BINDING CASSETTE SUB-FAMILY C |
| PE_Scaf9736_3.6 | |
| PE_Scaf9736_3.7 | GUANYLATE CYCLASE SOLUBLE SUBUNIT BETA-2 |
| PE_Scaf9736_3.8 | ATP-BINDING CASSETTE SUB-FAMILY C |
| PE_Scaf9736_3.9 | |
| PE_Scaf9736_5.4 | DESUMOYLATING ISOPEPTIDASE |
| PE_Scaf9736_5.6 | THYMIDINE PHOSPHORYLASE |
| PE_Scaf9736_6.7 | |
| PE_Scaf9736_6.8 | ZINC PHOSPHODIESTERASE ELAC PROTEIN 2 |
| PE_Scaf9739_0.0 | NADH-UBIQUINONE OXIDOREDUCTASE CHAIN 5 |
| PE_Scaf9739_0.1 | |
| PE_Scaf9739_0.3 | CYTOCHROME C OXIDASE SUBUNIT III |
| PE_Scaf9740_0.3 | BTK-BINDING PROTEIN-RELATED |
| PE_Scaf9740_1.1 | UNCHARACTERIZED |
| PE_Scaf9740_3.7 | PROTEIN DISULFIDE ISOMERASE |
| PE_Scaf9740_5.5 | |
| PE_Scaf9740_5.8 | |
| PE_Scaf9743_0.12 | ATP-BINDING CASSETTE SUB-FAMILY B |
| PE_Scaf9743_0.13 | ATP-DEPENDENT PERMEASE MDL1, MITOCHONDRIAL |
| PE_Scaf9744_0.7 | ARGININE DEMETHYLASE AND LYSYL-HYDROXYLASE JMJD |
| PE_Scaf9744_0.8 | SODIUM/CHLORIDE DEPENDENT TRANSPORTER |
| PE_Scaf9744_0.9 | SODIUM/CHLORIDE DEPENDENT TRANSPORTER |
| PE_Scaf9745_0.0 | |
| PE_Scaf9746_0.3 | |
| PE_Scaf9746_1.1 | |
| PE_Scaf9746_1.5 | ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE ALKB HOMOLOG 2 |
| PE_Scaf9746_3.4 | GUANYLYL CYCLASE DOMAIN CONTAINING PROTEIN 1 GUCD1 |
| PE_Scaf9746_3.5 | CT120 PROTEIN |
| PE_Scaf9746_4.6 | |
| PE_Scaf9746_5.0 | |
| PE_Scaf9746_6.4 | ANKYRIN REPEAT DOMAIN-CONTAINING PROTEIN 13 |
| PE_Scaf9746_6.5 | G PROTEIN-COUPLED RECEPTOR KINASE INTERACTING ARFGAP |
| PE_Scaf9746_6.6 | COLLAGEN IV NC1 DOMAIN-CONTAINING PROTEIN |
| PE_Scaf9748_0.3 | |
| PE_Scaf9748_1.6 | |
| PE_Scaf9748_1.7 | |
| PE_Scaf974_0.3 | DNA REPLICATION LICENSING FACTOR MCM FAMILY MEMBER |

