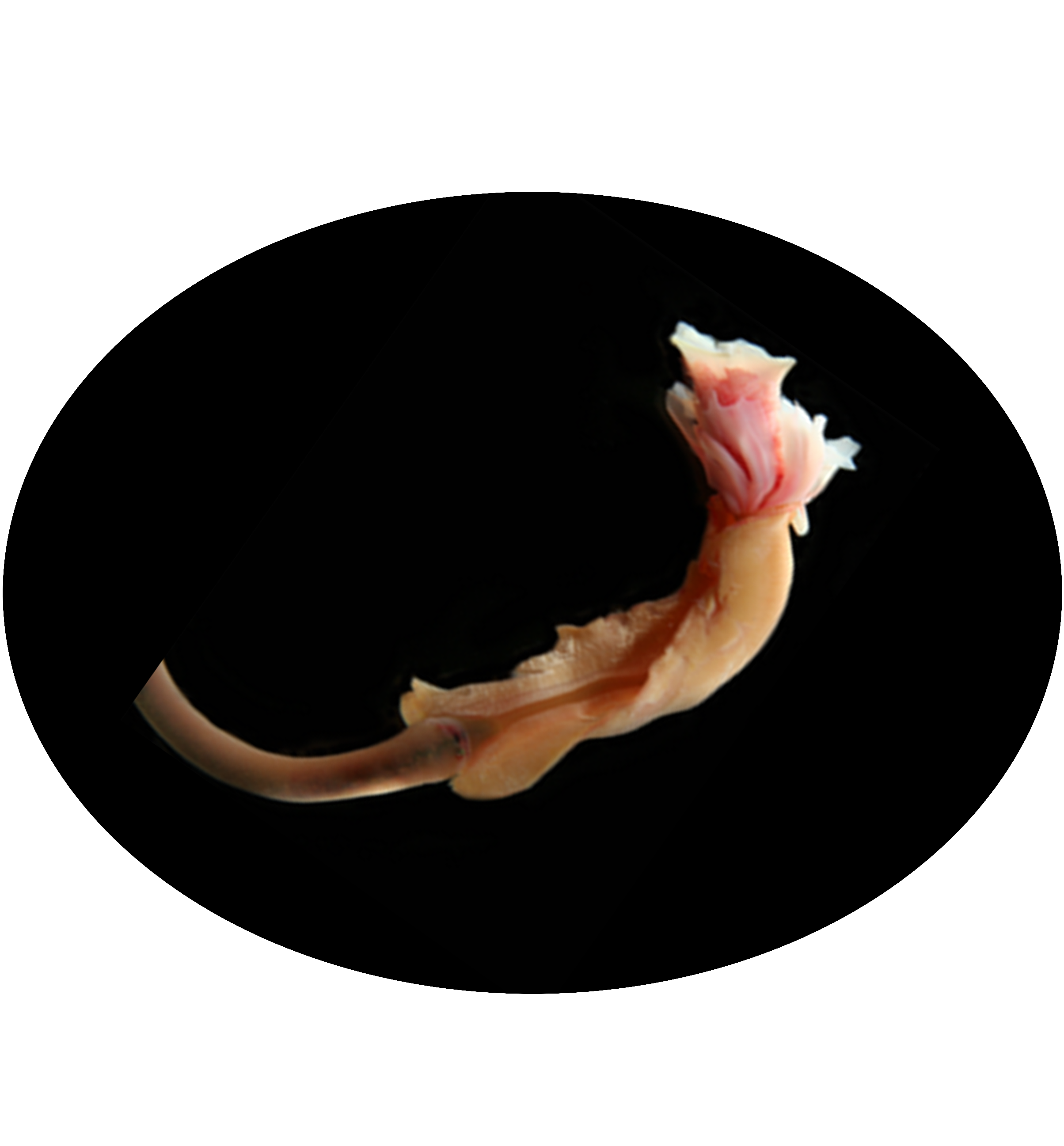
Lamellibrachia columna is a species of tube worm in the family Siboglinidae, found in deep-sea hydrothermal vents and cold seep ecosystems. This species thrives in extreme environments by relying on a symbiotic relationship with sulfide-oxidizing bacteria housed in its trophosome, a specialized organ. These bacteria convert hydrogen sulfide into organic compounds, providing the worm with essential nutrients. Lamellibrachia columna plays a vital role in deep-sea ecosystems by contributing to nutrient cycling and supporting diverse microbial and faunal communities.
Animalia (Kingdom); Annelida (Phylum); Polychaeta (Class); Sedentaria (Subclass); Canalipalpata (Infraclass); Sabellida (Order); Siboglinidae (Family); Lamellibrachia (Genus); Lamellibrachia columna (Species)
Lamellibrachia columna Southward, 1991
1. Southward E C. Three new species of Pogonophora, including two vestimentiferans, from hydrothermal sites in the Lau Back-arc Basin (Southwest Pacific Ocean)[J]. Journal of Natural History, 1991, 25(4): 859-881. (Southward et al., 1991)
2. Southward E C. Three new species of Pogonophora, including two vestimentiferans, from hydrothermal sites in the Lau Back-arc Basin (Southwest Pacific Ocean)[J]. Journal of Natural History, 1991, 25(4): 859-881. (Kobayashi et al., 2015)
Tube length 277.0–661.5 mm (mean=545.7 mm, n=4); outer width of top funnel opening 9.5–11.2 mm (n=4); width of basal end 2.8–7.8 mm (mean=4.5 mm, n=4). All tubes incomplete, lacking considerable parts of basal regions. External characters of tube variable along its length.Anterior part straight or slightly curved, but not coiled, with many short collars. Inter-collar distance generally small but varying among specimens. Posterior part sinuous, curled, smooth, without collars.
The specific epithet sagami,asnoun in apposition, refers to the province name of the Edo period for Kanagawa, the coastal area of Sagami Bay, the type locality
The species is known from cold seep areas off Hatsushima and on the Okinoyama Bank, Sagami Bay, the Kanesu-no-se Bank, the Ryuyo Canyon, the Omaezaki Spur and the Tenryu Knoll, the Nankai Trough, and the Kuroshima Knoll, the Ryukyu Trench between 270–1300 m. It is also known from hydrothermal vent fields on the Iheya Ridge and the North Iheya Knoll, the Okinawa Trough, and the Sumisu Caldera, the Izu-Bonin arc between 900–1500 m depth
| Species | Phylum | Common Name | Ecosystem | Depth | Habitat | NCBI Taxonomy ID |
|---|---|---|---|---|---|---|
| Lamellibrachia columna | Annelida | - | Cold seep | 810-1,870 | cold seeps off the east coast of the North Island of New Zealand | 53616 |
| Genome Assembly | Genome Size | Assembly level | Released year | WGS accession | Submitter | BioProject | BUSCO completeness (%) | Scaffold/Contig N50 (kb) | GC content (%) | Repeat Rate (%) | Gene Number |
|---|---|---|---|---|---|---|---|---|---|---|---|
| wsLamColu1.1 | 879.7Mb | Chromosome | 2023 | CAVLEN01 | WELLCOME SANGER INSTITUTE | PRJEB68353 | 97.30 | 472.9/472.5 | 40.5 | - | 32,318 |

